- Department of Biological Sciences
- Campus Experiences
- Facilities
- Molecular Core Lab
Molecular Core Lab
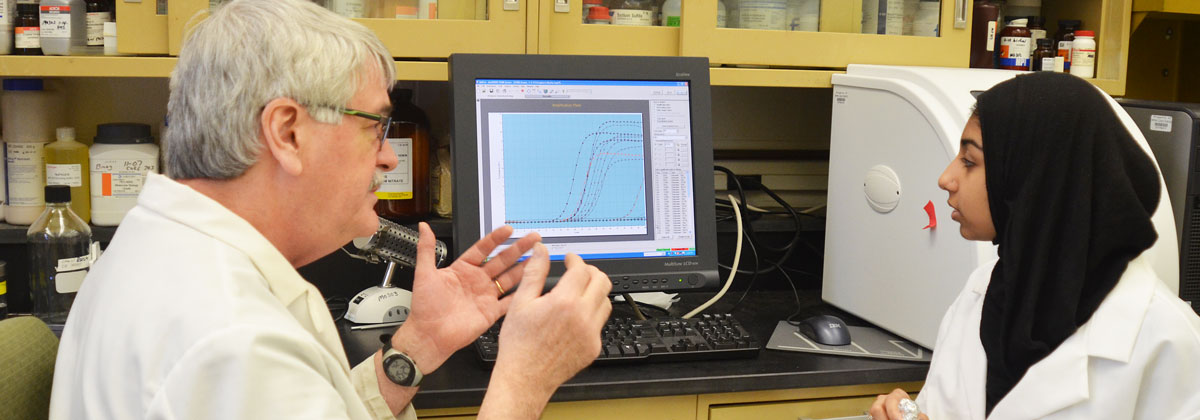
The main focus of the Molecular Core Lab is support for purification of DNA and RNA and the use of molecular biology equipment for analysis of gene expression and genotyping. Dr. Grayburn has been working in labs with nucleic acids since 1981 and can help students with many aspects of molecular biology. Centrifuges, thermal cyclers, lyophilizers, autoclaves, ovens, a dishwasher, and gel documentation equipment are available for student use.
Gene expression studies frequently use quantitative real-time PCR (qPCR). This procedure has been used to determine if specific genes are expressed at different levels in response to growth conditions or stage of developmental. It can also be used to investigate how a gene deletion affects the expression of other genes. Many studies in different organisms are possible with this technology. A Mx3000P instrument from Agilent Technologies is available to perform these reactions.
The Molecular Core Lab uses capillary electrophoresis for DNA genotyping analysis. It is possible to distinguish individual plants, snakes, or other creatures from each other. In this assay, PCR products are labeled with fluorescent dyes that can be detected using a Beckman Coulter CEQ 8000 Genetic analysis system.
Undergraduate Research
Opportunities are available for undergraduate students to obtain hands-on research in a molecular biology lab. Dr. Grayburn has several projects that are available for undergraduate participation. One research area is renewable biofuel from algae cellulose. Cellulose is a polymer of glucose (sugar) molecules which is often the dominant structural molecule in algal cell walls. As seen with cotton clothing, which is made of cellulose fibers, it is durable and difficult to hydrolyze into simple sugars. Expression of genes involved in this process are being studied in a locally-isolated bacterium.
Another research area studies viruses of freshwater algae. Viruses are the most abundant life form on earth, but the majority of viruses have not been studied. Many times the DNA or RNA sequence of these viruses differs from information in public databases, making identification of these organisms difficult. An established model system is being used to develop techniques for enrichment of algae viruses from local water bodies.

Scott Grayburn
Contact: sgrayburn@niu.edu
(815) 753-0638
Office: MO 344
Lab: MO 303